It is really a while that I search for a way to store diverse data, most of them grids, in a repository where the metadat would be preserved and in case even editable. A place where one could extract subsets of data from the datasets. A place where one could access the data also directly from the GIS... yes, I know, everything is thinking about OGC and some WCS and whatever else.
But the summer of code opened me a pandora box full of presents and by choosing the necdf format many possibilities built on open sourced software are available.
Ramadda can be tested on the unidata's demo server, so I won't talk about the many features that one can try out there.
I want to talk about background jobs that can be done from the client code of Ramadda, which Mr. Jeff McWhirter was so kind to introduce me to and help me with.
First, to get started, you need the repository client library, which you can download here. Once the contained libraries are in the classpath, you can start.
Let us assume we start from an existing ramadda repository, that looks like the following:
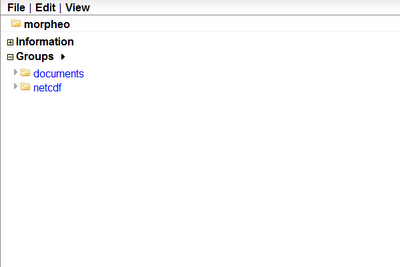
Let's assume we need to upload a netcdf file that has been produced in JGrass by some model.
The following shows the code needed to do so.
First a client connection has to be instantiated:
RepositoryClient repositoryClient = new RepositoryClient(host, Integer.parseInt(port), base, user, pass);
String[] msg = {""};
if (!repositoryClient.isValidSession(true, msg)) {
// throw some exception
}
where the isValidSession method in this case also performs a login in the ramadda environment.
Once the connection is done, the upload of any file can be done. The following method does that for you:
/**
* Uploads a file to ramadda.
*
* @param entryName name of the entry that will appear in the ramadda server.
* @param entryDescription a description of the entry that will appear in the ramadda server.
* @param type the file type that is uploaded.
* @param parent the parent path inside the repository, into which to upload the file
* ex. morpheo/documents, where morpheo is the base group and documents
* the subgroup.
* @param filePath the path to the file to upload.
* @throws Exception
*/
public String uploadFile( String entryName, String entryDescription, String type,
String parent, String filePath ) throws Exception {
Document doc = XmlUtil.makeDocument();
Element root = XmlUtil.create(doc, TAG_ENTRIES, null, new String[]{});
Element entryNode = XmlUtil.create(doc, TAG_ENTRY, root, new String[]{});
/*
* name
*/
entryNode.setAttribute(ATTR_NAME, entryName);
/*
* description
*/
Element descNode = XmlUtil.create(doc, TAG_DESCRIPTION, entryNode);
descNode.appendChild(XmlUtil.makeCDataNode(doc, entryDescription, false));
/*
* type
*/
if (type != null) {
entryNode.setAttribute(ATTR_TYPE, type);
}
/*
* parent
*/
entryNode.setAttribute(ATTR_PARENT, parent);
/*
* file
*/
File file = new File(filePath);
entryNode.setAttribute(ATTR_FILE, IOUtil.getFileTail(filePath));
/*
* addmetadata
*/
entryNode.setAttribute(ATTR_ADDMETADATA, "true");
ByteArrayOutputStream bos = null;
ZipOutputStream zos = null;
try {
bos = new ByteArrayOutputStream();
zos = new ZipOutputStream(bos);
/*
* write the xml definition into the zip file
*/
String xml = XmlUtil.toString(root);
zos.putNextEntry(new ZipEntry("entries.xml"));
byte[] bytes = xml.getBytes();
zos.write(bytes, 0, bytes.length);
zos.closeEntry();
/*
* add also the file
*/
String file2string = file.toString();
zos.putNextEntry(new ZipEntry(IOUtil.getFileTail(file2string)));
bytes = IOUtil.readBytes(new FileInputStream(file));
zos.write(bytes, 0, bytes.length);
zos.closeEntry();
} finally {
zos.close();
bos.close();
}
List postEntries = new ArrayList();
postEntries.add(HttpFormEntry.hidden(ARG_SESSIONID, repositoryClient.getSessionId()));
postEntries.add(HttpFormEntry.hidden(ARG_RESPONSE, RESPONSE_XML));
postEntries.add(new HttpFormEntry(ARG_FILE, "entries.zip", bos.toByteArray()));
RequestUrl URL_ENTRY_XMLCREATE = new RequestUrl(repositoryClient, "/entry/xmlcreate");
String[] result = repositoryClient.doPost(URL_ENTRY_XMLCREATE, postEntries);
if (result[0] != null) {
outputStream.println("Error:" + result[0]);
return null;
}
Element response = XmlUtil.getRoot(result[1]);
String body = XmlUtil.getChildText(response).trim();
if (repositoryClient.responseOk(response)) {
outputStream.println("OK:" + body);
} else {
outputStream.println("Error:" + body);
}
Element child = XmlUtil.findChild(response, "entry");
String entryId = child.getAttribute("id");
String urlString = "http://" + host + ":" + port + base + "/entry/get/" + entryName
+ "?entryid=" + entryId;
return urlString;
}
This method returns a url string, that can be used in the browser to fetch the uploaded dataset.
So if I was to upload a netcdf called water_surf.nc without supplying a particular path, it would have resulted in appearing in the base group, called morpheo in this case:
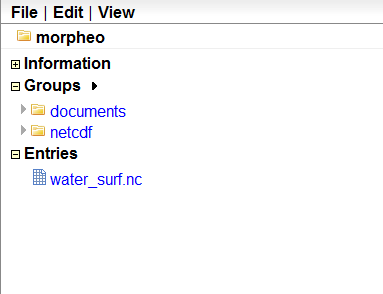
What I really love about ramadda, is that if the dataset is in netcdf format for example, the metadata are accessible and editable also from the web interface:
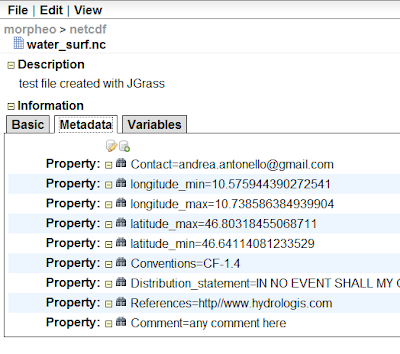
There are several ways the data can be accesses, and note in the below picture the opendap link, which is the one the JGrass uses to visualize the dataset or use it in the models:
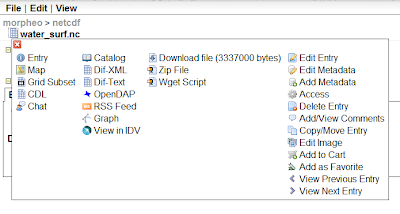
Ok, but I was writing about programmatically access the data. So how to fetch data from the server? They can be downloaded from ramadda by means of their id. In fact the last part of the above returned url string represents the unique id of the dataset (...?entryid=THEENTRYID).
Downloading a file from ramadda is incredibly easy, since the RepositoryClient class sopplies a method called writeFile that takes the entry id and the output file to which to download as parameters:
repositoryClient.writeFile(entryId, outputFile)
And that is all, thanks to Jeff McWhirter for all the great help he gave me.
Soon I will be glad to describe some deeper integration between JGrass modeling environment and Ramadda.
1 comment:
Great post! It was very useful ;)
Thanks!
Regards
Post a Comment