It is some time now that the
Geo-solutions guys created at geotools level tools for tiling, mosaicing and creating overviews of images.
It was really time to have them in udig to use them on those images that trouble us with memory overload.
I have thought a thousand of times about creating a wizard at Udig level, so it could be in the standard Udig, but in the end I brought it as OpenMI based module into JGrass for several small reasons, but one main reason.
As JGrass module it is scriptable and it also gets almost automagically a gui. Also the scripts can be launched with the console in standalone mode on serverside. So after a small discussion with Simone of Geosolutions we decided for them to be a JGrass module.
That means that to use those for now you will have to load the JGrass plugins.
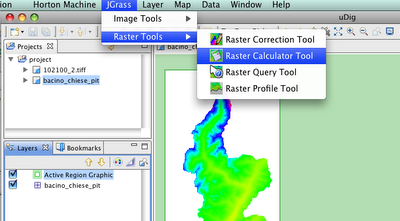
So, what are those coverage tools?Imagine a schenario of the following kind: you have 25 geotiffs, that take one hour to load the first 3 of them and than freezes udig and at some point breakes completely.
This is probably due to the fact that those geotiffs do not have overviews.
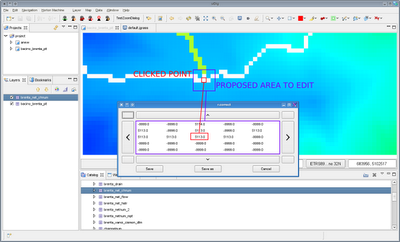
Now you can solve it from Udig with the following:
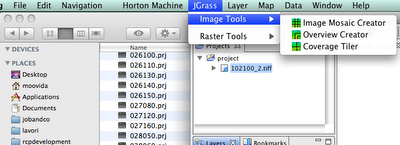
Choose the Overview creator and you will get a dialog:
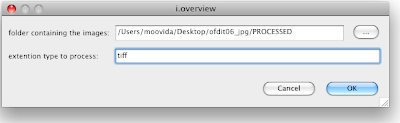
enter the folder and the extention of your images and press ok.
The overview tool takes a hell of a time and for that hell of a time on my pc it takes all resources. And I mean ALL. Launch it best before going to sleep.
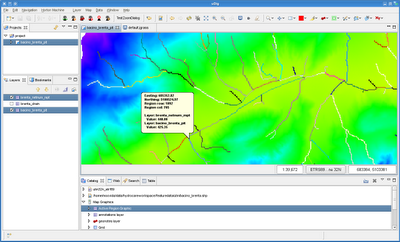
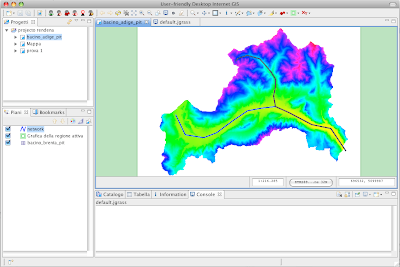
After the creation of overviews, your geotiffs should load really fast and without sucking your pc memory. Now I get really quickly my images loaded:
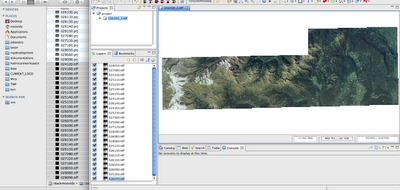
Well, what we do not like, is that we have 25 layers with one image each.
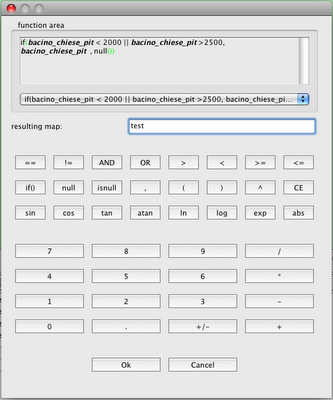
That is why in the coveragetools there is also an
Image Mosaic Creator. Launch it:
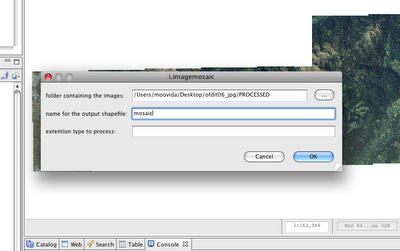
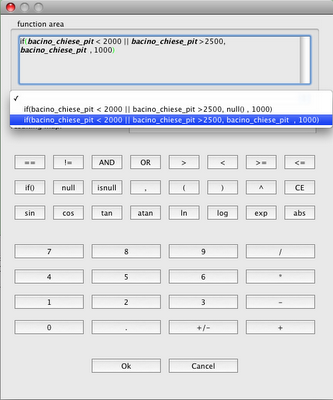
Since the extension defaults to tiff, I can omit it.
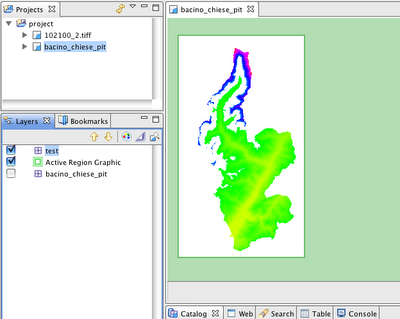
After a few seconds the shapefile is created and if I load it, the result is the following:

Two main differences:
1) the layer is just the one of the shapefile that loads all the geotiffs (great!!)
2) the background of missing tiles is black (ugly)
There is also a third tool, the Image tiler, which splits an image into smaller pieces of the dimension the user supplies.
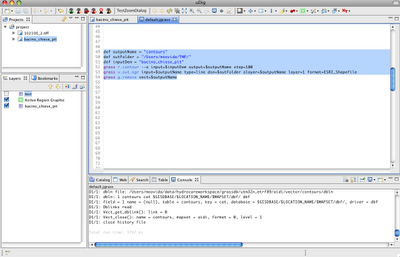
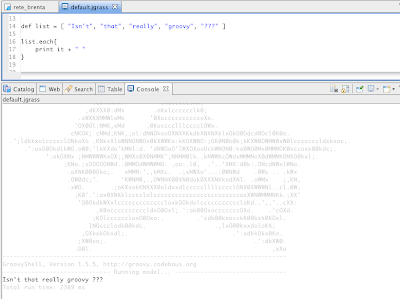
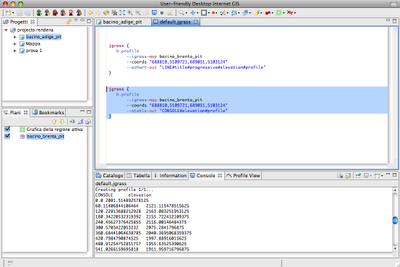
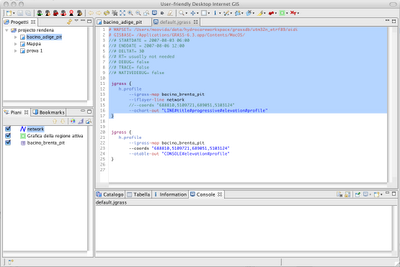
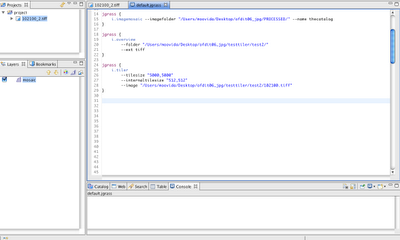
Alright, as promised for a bit more advanced users the same commands can be accessed from the console. In that mode it will be also possible to tweak some flags and values, which in the gui are set to default.
The scripts look like that:
This is really a nice set of tools for all those that do not have much memory.
The downside of the tools is that they work with the new geotools and therefore only with the new development version. But hey, in Cape Town we will try to use that one, so you will be able to get your Udig with all the tools in it by next month.